The reversible-binding systems mentioned in the previous post are examples of non-specific capture methods. They capture total DNA and RNA in a sample because they simply rely on the affinity of nucleic acids to the magnetic particle coating or functional moiety. Non-specific capture methods bind all single-strand (ssDNA) or RNA regardless of sequence. A more specific capture system is able to target specific sequences of ssDNA or RNA. This type of specific capture is most applicable to diagnostic systems as an assay for a specific pathogen. Examples include systems to diagnose methicillin resistant staphyloccocus aureaus (MRSA), specific plant viruses, and infection with malaria-causing bacteria. These strategies could be applied to detect viral, bacterial, or fungal pathogens in a variety of clinical samples.
Free donwload: Magnetic DNA purification
One method for specific capture of single strand nucleic acids involves the attachment of oligonucleotide probes onto the surfaces of magnetic particles. The oligonucleotide probe is fully customizable and is synthesized to be complementary to the target sequence. There are a couple of ways to attach the probes to the surface of the particles. In one system the single-strand oligonucleotides are attached with a non-cleavable triethylene glycol spacer. Another method of capturing specific ssDNA or RNA sequences relies on streptavidin-biotin affinity. Streptavidin and biotin have a very strong specific affinity for each other, and this is a common system for attaching moieties to magnetic particles. The magnetic particles are coated with streptavidin molecules and can bind biotin-tagged single-strand oligonucleotide probes during an incubation period. The biotin tags are added during the initial synthesis of the probes.
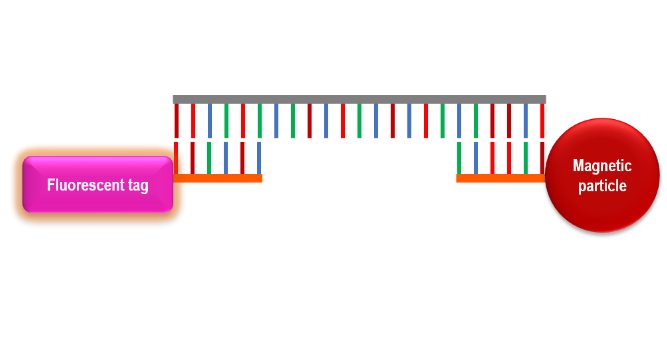
Specific nucleic acid capture is highly valuable in situations where the isolation of a specific sequence of DNA or RNA is desired. This type of magnetic particle is very useful in real-time DNA/RNA detection systems if it is combined with a fluorescent or chemiluminescent probe. The simultaneous binding of an oligonucleotide probe and a detection probe to a target ssDNA or RNA strand is called a sandwich hybridization immunoassay.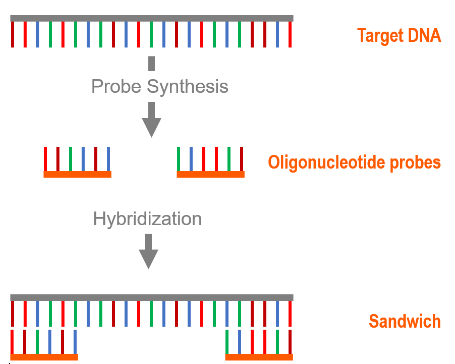
Sandwich hybridization is a commonly used method in specific ssDNA or RNA capture for diagnostic purposes. Sandwich hybridization involves two oligonucleotide probes that bind to either end of the target ssDNA or RNA. One probe is synthesized complementary to a section of the 5' to 3' end, and the other is complementary to a section of the 3' to 5' end. One probe is conjugated to a magnetic particle. The other probe is conjugated to a diagnostic molecule. When the target ssDNA or RNA is present, the two probes bind to it and form a single complex.
Sandwich hybridization provides a real-time quantitative or qualitative read-out of capture efficiency. The diagnostic molecule depends on whether the read-out is based on a system using fluorescent probes and quantum dots for simple fluorescent signal readout, a more sophisticated fluorescence resonance energy transfer (FRET), molecules for surface-enhanced Raman spectroscopy (SERS), or molecules for a chemiluminescent reaction. A chemiluminescent reaction is a chemical reaction that produces a high-energy intermediate that emits a photon of light as it reacts to its final product. The ability to specifically bind a target sequence of ssDNA or RNA is powerful for identifying the presence or absence of a pathogen. When it is combined with a light-emitting probe this technology becomes extremely valuable as a point-of-care diagnostic tool.
Related news